RNA Secondary Structure
Problem1
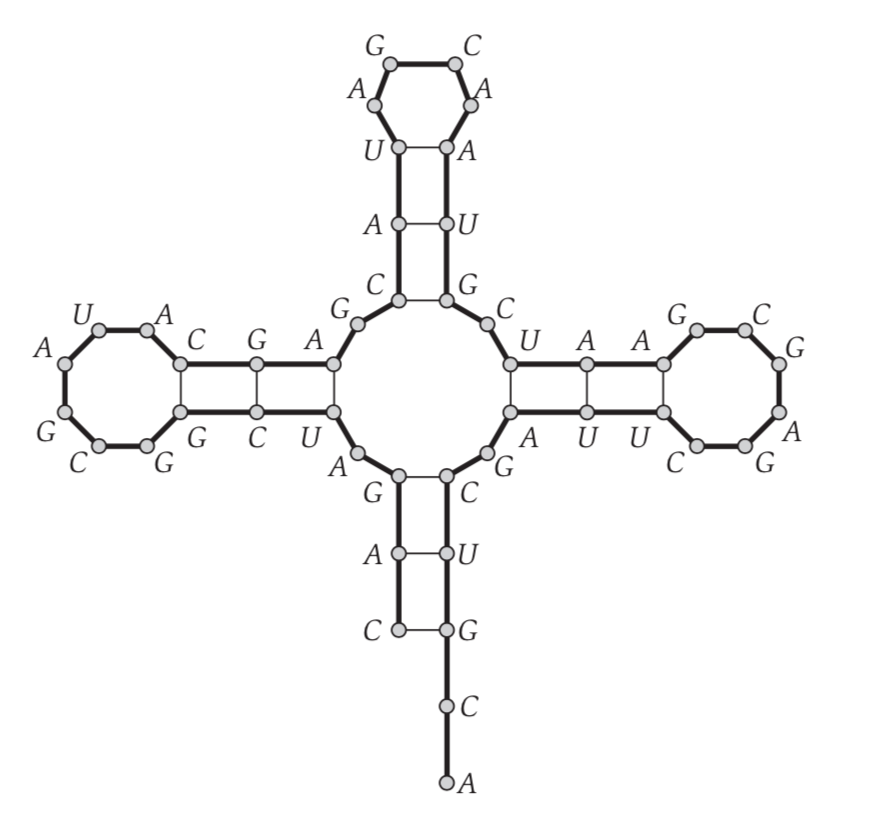
We have bases of {A,C,G,T} in DNA sequence, where A-T and C-G form a pair. A sinlge strand of RNA will loop back, resulting shape called secondary structure.
Let $B = b_1b_2 … b_n$, where $b_i$ = {A,C,G,U} also A-U, C-G form pair
Then the Secondary structure, S ={(i, j)}
- no sharp turn, i < j - 4
- pair is either {A,U}, {C,G} (either order)
- no base appear in more than 1 matching
- no crossing condition, (i,j) and (k, l) $\in$ S, we can’t have i < k < j < l
Algorithm
Following the same analysis
- OPT(j) = the maximum number of base pairs in a secondary structure on $b_1b_2 … b_j$.
- OPT(j) = 0 for j $\leq$ 5
The above rules (constraints) make the recurrent relationship
- j is not involved in a pair
- j pairs with t for some t < j − 4
Then we split the problem into two cases due to noncrosing constriant (no pair form between first part and second part)
- from 1 to t - 1 using OPT(t − 1)
- from t + 1 to j - 1 which is not considered by our OPT
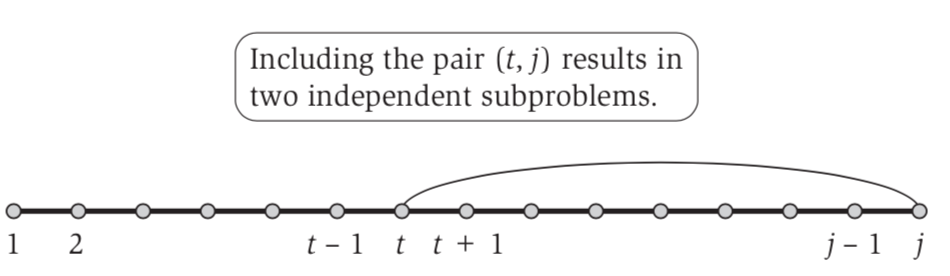
This push our OPT take take another paramter i, so OPT(i,j) = maximum number of base pairs in a secondary structure on $b_ib_{i+1} … b_j$.
Therefore, OPT(i, j) = max(OPT(i, j − 1), max(1 + OPT(i, t − 1) + OPT(t + 1, j − 1))) (6.13).
- Initialize OPT(i,j)=0 whenever i≥j−4
- For k=5, 6,…,n−1
- For i=1,2,…n−k
- Set j=i+k
- Compute OPT(i, j) using the recurrence in (6.13)
- Endfor
- For i=1,2,…n−k
- Endfor Return OPT(1, n)
Analysis
There are $O(n^2)$ subproblems to solve, and evaluating the recurrence in (6.13) takes time O(n) for each. So overall $O(n^3)$.
- Algorithm Design by Jon Kleinber, Eva Tardos [return]